terpenoid Summary
Terpenoid are key components of human medicines and are the largest family of natural products. Fungi are important sources of terpenoids, but many of the corresponding biosynthetic gene clusters (BGCs) are silent under laboratory conditions.

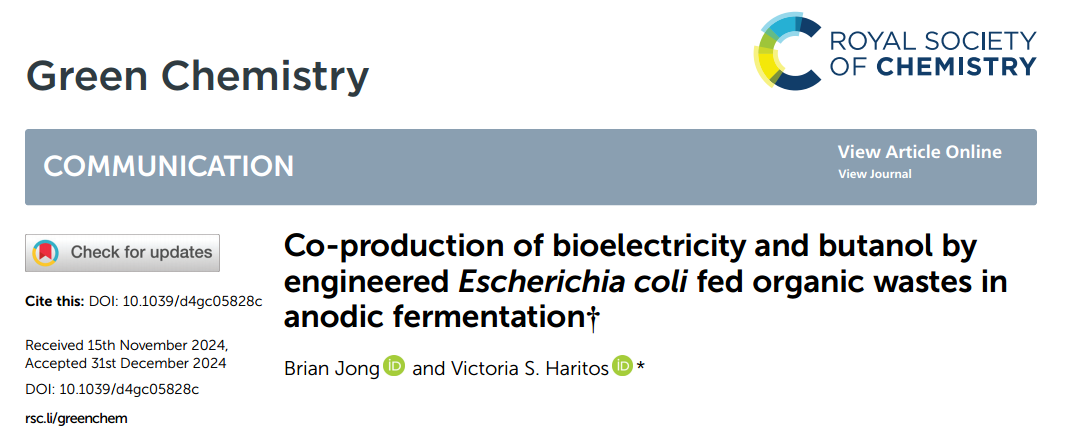
Strategies such as homologous activation and heterologous expression often used to activate a single cluster and inefficient. Therefore, using Aspergillus oryzae as the basis, the researchers developed an automated and high-throughput (auto-HTP) biobased workflow that can effectively perform genome mining, characterization of BGCs, and identification of bioactive fungal terpenoids. With the help of this platform, researchers completed the recombination of 39 BGCs, obtained 208 engineering strains, and produced 185 different terpenoid compounds.
The sesquiterpene mangicol J discovered through an anti-inflammatory screen; retrospective studies on engineered strains revealed possible biosynthetic pathways.
Finally, the researchers optimized the mevalonate pathway in Aspergillus oryzae, providing a more efficient basis for the overproduction of terpenoids. Automated high-throughput work platform processes and optimized Aspergillus oryzae chassis can effectively promote the discovery and development of terpenoid natural products.
main content
1. Bioinformatics analysis and reconstruction principles of terpenoid BGCs
The filamentous fungus Aspergillus oryzae (AO) has a cellular environment more conducive to the exploration of fungal natural products and is an ideal chassis for mining individual terpene cyclases or entire BGCs from filamentous fungi via CRISPR–Cas9.
However, the limited supply of precursors (e.g., isopentenyl pyrophosphate and dimethylallyl pyrophosphate) makes it difficult to obtain sufficient terpenoids for further biochemical and functional studies, especially for those that require multiple Late derivatives of biosynthetic steps.
Therefore, the researchers selected AO NSAR1 as the v.1.0 chassis strain and developed an automated and high-throughput (auto-HTP) biological workflow as a new genome mining strategy.
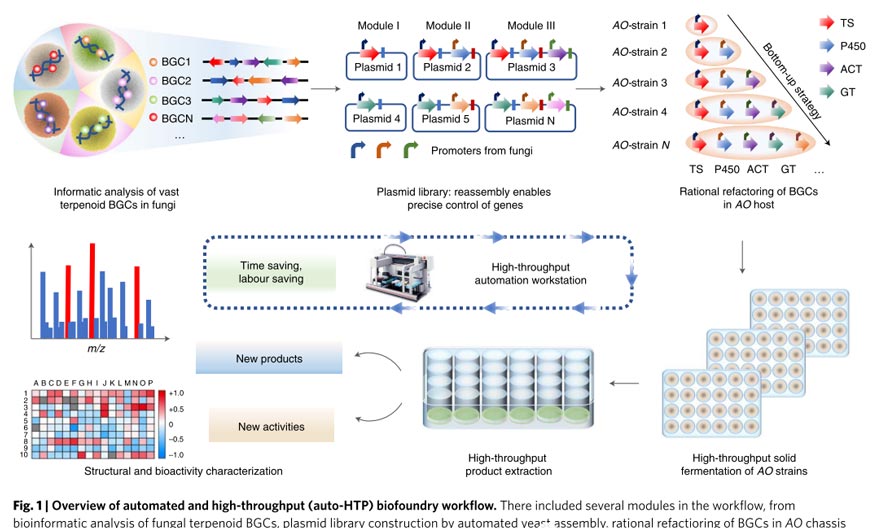
The specific strategic principles shown in the figure: antiSMASH used to predict BGCs from five internally sequenced filamentous fungi, and domains that do not contain known conserved domains or have incomplete functions deleted.
Then the BGCs divided into three modules: upstream terpene synthase, midstream P450 oxidation and downstream modification for assembly, and the functional genes placed under pre-characterized strong constitutive (hlyA) and inducible (amyB, glaA) promoters.
With the help of automated high-throughput work platforms, genes from the same module placed in separate plasmids or co-constructed into one plasmid for all possible combinations.
Based on the combination of plasmids from different modules, the terpene synthase gene used as the basic starting point, and then other downstream genes from the same BGC added respectively, thereby reconstructing BGCs into a strain library.
2. Automated and high-throughput workflows
With the help of automation and high-throughput biological workflows, the researchers combined 39 BGC reconstructions and obtained 166 (96%) plasmids and 195 (96%) AO strains after two rounds of construction.
The remaining 7 plasmids and 13 strains contained multiple genes and large plasmids respectively and artificially constructed. The entire process from PCR amplification to final construction of the AO strain takes 17 days.
Finally, the engineered AO strain incubated in 24 deep-well plates containing solid rice medium at 30°C for 2 weeks, and compounds extracted from the culture twice using acetone and ethyl acetate and concentrated to produce a crude extract. things.
Using GC–MS and HR–ESI–MS, 185 different terpenoids detected in 208 AO strains, including 23 sesquiterpenes, 59 diterpenes, and 103 sesquiterpenes (22 of which sesquiterpenes). reported compound).
3. Screening of terpenoids with anti-inflammatory activity
The researchers treated RAW 264.7 mouse macrophages with lipopolysaccharide (LPS) to initiate inflammatory pathways and used a nitric oxide assay kit to monitor the release of nitric oxide (NO) as a readout; the ability of the crude extract to reduce NO levels was as a measure of its anti-inflammatory activity.
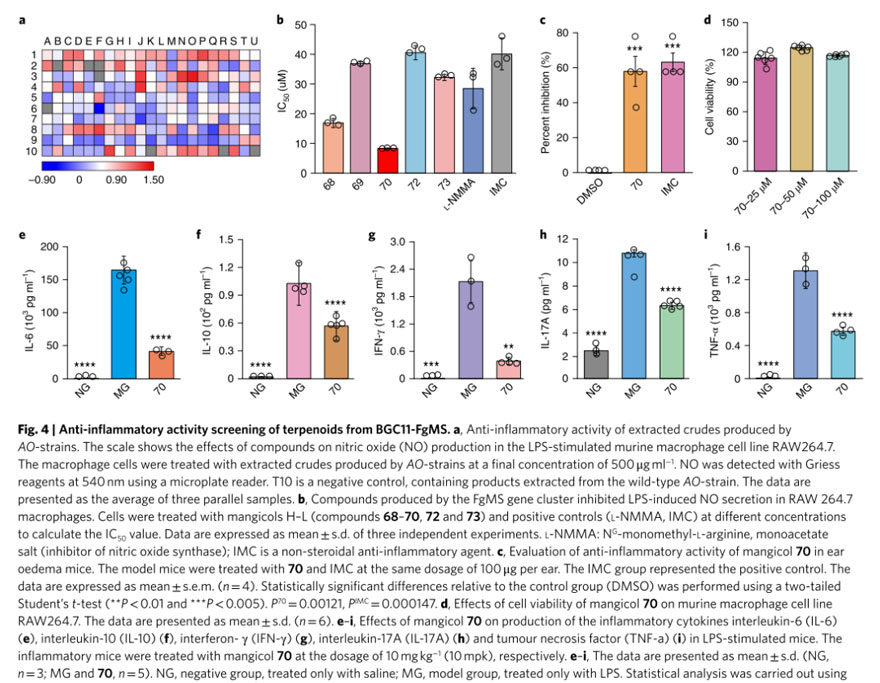
Testing with the help of an automated high-throughput biological work platform showed that 16% of the fermentation extract showed significant anti-inflammatory activity and could reduce NO levels to less than 20% of the untreated control. Among them, strains AO-Y52 and Y5658 showed particularly high activity. These strains both contain genes from FgMS-BGC, which predicted to synthesize the reported anti-inflammatory sesquiterpenoids mangicols.
The researchers then traced all strains containing these genes, AO-Y51 to AO-Y62, and purified the corresponding compounds 68–73 and characterized them by NMR spectroscopy. Compound 71 identified as the known molecule mangicol E; compounds 68–70, 72, and 73 named new sesquiterpenoids mangicol H–L, respectively.
In vitro data showed that mangicol J had the highest NO inhibitory activity and was more effective than the positive controls L-NMMA (nitric oxide synthase inhibitor) and IMC (nonsteroidal anti-inflammatory agent) without cytotoxicity.
On this basis, the researchers used a mouse ear edema model to evaluate the anti-inflammatory activity of mangicol J in vivo and found that it inhibited PMA-induced edema to the same level as the drug IMC.
The researchers also used the LPS-stimulated C57BL/6J mouse model and found that 10 mpk mangicol J significantly reduced serum levels of IL-6, IL-10, IFN-γ, IL-17A, and TNF-α.
Western blot data also showed that mangicol J could reduce the expression levels of phosphorylated signal transducer and activator of transcription 3 downstream of IL-6 in human umbilical vein endothelial cells.
4. Analyze the biosynthetic pathway of mangicol J and construct AO 2.0 chassis cells
An advantage of the automated HTP biobased workflow is that the reconstructed gene clusters enable rapid assessment of specific genes required for compounds of interest.
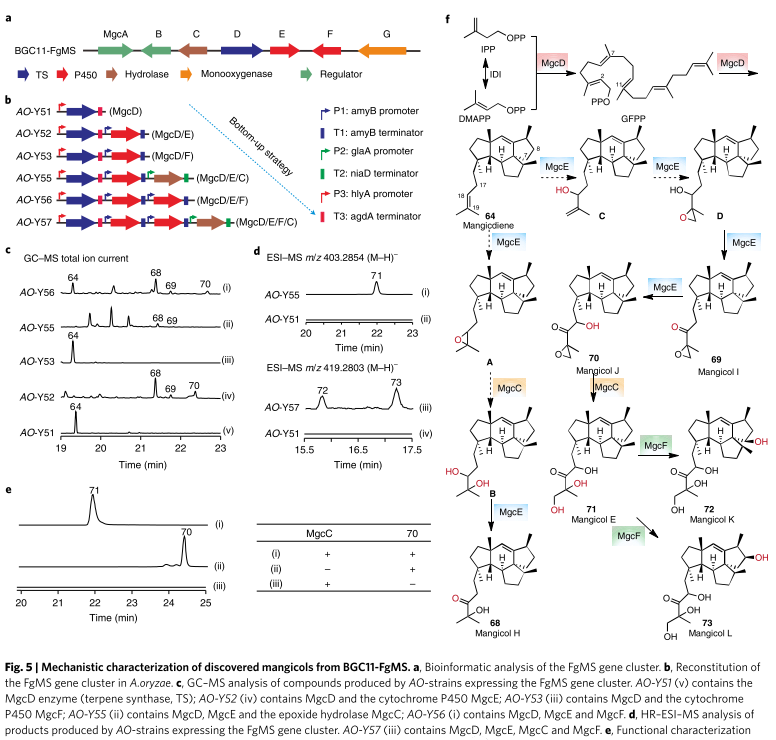
As mentioned above, strains that can produce mangicol J all include genes from the FgMS cluster. MgcD (FgMS) known to be a sesquiterpene cyclase that catalyzes the formation of 64 (mangicol). In AO-Y52 (containing MgcDE Mangicol H–J identified in ) deduced that MgcE is a multifunctional P450 capable of catalyzing the hydroxylation and epoxidation of the isoprenyl tail (C17–C20) of mangodiene;
Mangicol E detected in AO-Y55 (containing MgcCDE). It inferred that mgcC can catalyze the ring-opening hydrolysis of C19, C20 epoxides of Mangicol J and an epoxide hydrolase; detected in AO-Y52 (containing MgcDE) Mangicol H deduced that the natural epoxide hydrolase of AO can catalyze the ring opening from A to B, and finally synthesize 68;
Mangicol K and L derived from Mangicol E via C7 and C8 hydroxylation, respectively, were detected in strain AO-Y57 (containing MgcCDEF), indicating that P450 mgcF acts on the core structure. Combining these results, the putative biosynthetic pathway of mangiferol H–L was successfully predicted.
To scale up the production of the compounds
the researchers systematically engineered the MVA pathway of AO to build an efficient Chassis 2.0 for overproduction of mangicdiene and mangicol J.
First, a CRISPR–Cas9-mediated site-specific integration system was developed based on the previously developed CRISPR–Cas9 method. Strains AO-S81 to AO-S84 were obtained by constructing plasmids containing the entire endogenous MVA pathway, three additional copies of tHMG1 and mgcD, and randomly inserting them into the chromosome of AO NSAR1.
AO-S95 was generated by integrating the entire MVA pathway and additionally integrating four copies of tHMG1 at different HS sites of AO. One copy of mgcD was then integrated into the HS801 site of AO-S95 to create AO-S96.
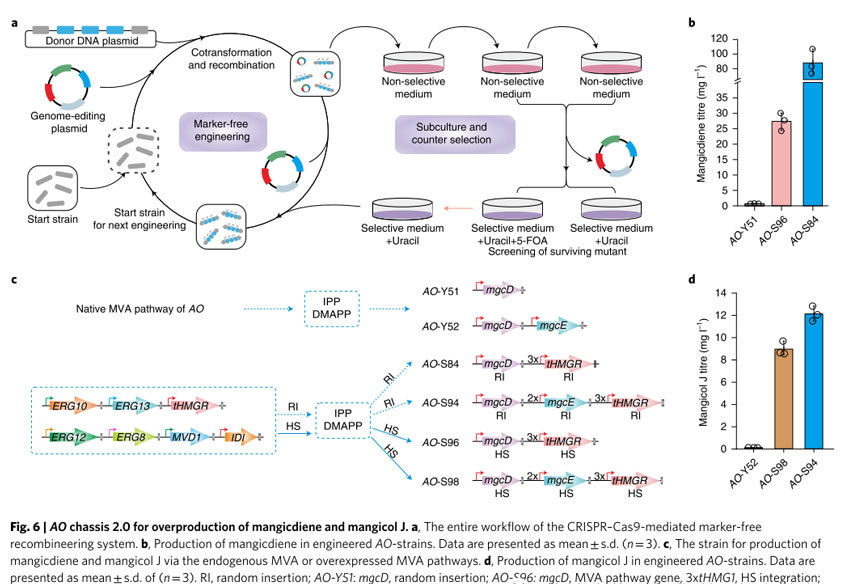
Compared with the parent strain AO-Y51, the titer of mangicdiene increased from 0.66 mg/L to 27.38 mg/L, a 41-fold increase in AO-S96, and to 87.84 mg/L in AO-S84, a 133-fold increase.
These titers are significantly higher than those of sesquiterpenes detected in E. coli and Bacillus cereus, providing an ideal platform for high-throughput genome mining of filamentous fungal-derived terpenoid BGCs.
Next, use random insertion to insert two copies of mgcE into the chromosome of AO-S84 to obtain strain AO-S94; use site-specific integration to integrate one copy of mgcD and two copies of mgcE into the chromosome of AO-S95. Strain AO-S97 obtained at the HS801 locus.
Compared with strain AO-Y52, the titer of mangicol J reached 8.93 mg/L in AO-S98, a 112-fold increase, and reached 12.09 mg/L in AO-S94, a 151-fold increase.
Finally, mgcD knocked out from strain AO-S84 to generate AO-S85 as a universal chassis for characterizing genes and terpenoid structures 2.0. The versatility of this advanced chassis confirmed by reconstituting the BGC37-Tvi09626 gene cluster into the AO-S95 strain. The titers of oxidation products and glycosylation products significantly higher compared to Chassis 1.0.
Summarize
In this study, the authors developed an automated and high-throughput (auto-HTP) biological workflow using Aspergillus oryzae as a chassis, enabling efficient genome mining, characterization of BGCs, and identification of bioactive fungal terpenoids.
At the same time, 39 BGCs recombined into 208 engineered strains, producing 185 different terpenoids. Anti-inflammatory screening identified the sesquiterpene mangicol J and revealed its possible biosynthetic pathway.
In addition, the mevalonate pathway in Aspergillus oryzae also optimized to obtain the AO 2.0 chassis, which provides a more effective basis for the mass production of terpenoids.