Systems engineering of Escherichia coli for highlevel shikimic acid

Research background
shikimic acid is a common precursor of drugs and industrial products, including the anti influenza drug Tamiflu (oseltamivir phosphate), aromatic compounds, and chiral chemicals.
So far, different host strains including Corynebacterium glutamicum, yeast, and Escherichia coli have engineered to increase shikimic acid production.
Among them, the highest production of shikimic acid titer achieved by inhibiting cell reactions through the growth of glutamic acid rod-shaped bacteria, with a concentration of 141 g/L. Among yeast strains, the highest shikimic acid titer found in Schillosomia stipites, with a concentration of 3.11 g/L.
However, the fermentation process of shikimic acid is complex and has low potency, which limits its industrial production. Escherichia coli has the advantages of simple operation, clear metabolic pathways, fast growth, and high carbon absorption rate, making it an attractive host bacterium for shikimic acid production.
research contents
In order to increase the shikimic acid production of tryptophan producing host strain E.coli TRP0, an enzyme constrained model ec was first used_ IML1515 has identified key gene targets that promote the biosynthesis of shikimic acid.
Then engineering these targets into E. coli TRP0 to produce shikimic acid. The accumulation of quinic acid and DHS byproducts significantly reduced through ydiB deficiency and AroE protein engineering, respectively.
1. Utilizing EC_ Identification of key targets for shikimic acid production using the iML1515 model
Using Escherichia coli as the substrate and glucose as the substrate, a total of 90 proteins screened from a simulated database, including 7 high demand proteins and 83 low demand proteins.
On this basis, 12 potential targets that directly affect the biosynthesis of shikimic acid selected from the screened proteins through metabolic pathways, literature review, and predicted flux sorting.
Due to the fact that shikimic acid an upstream product of tryptophan, a tryptophan producing strain E. coli TRP0 selected through random mutagenesis as the substrate for fermentation to produce shikimic acid.
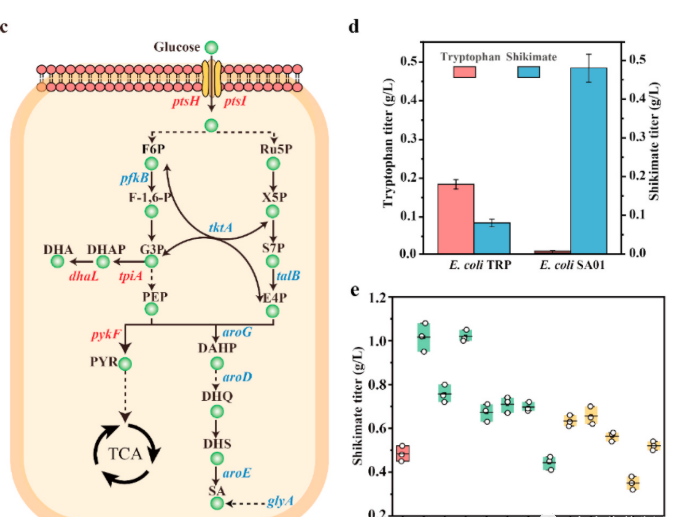
In order to accumulate the yield of shikimic acid in E.coli TRP0, both aroK (encoding shikimic acid kinase I) and aroL (encoding shikimic acid kinase II) are missing to block the conversion of shikimic acid to its downstream product tryptophan.
The obtained strain E.coli SA01 produced 479 mg/L of shikimic acid through shake flask fermentation. Meanwhile, the titer of its downstream product tryptophan significantly decreased to<10 mg/L.
These results indicate that the E.coli TRP0 strain successfully transformed to accumulate shikimic acid instead of tryptophan, and the obtained strain E.coli SA01 used as a parent strain for subsequent research.
2. Constructing Escherichia coli with excessive production of shikimic acid
Based on 10 effective target genes, we adopted a series of gene manipulation strategies to increase the titer of shikimic acid. Preliminary studies conducted on Escherichia coli SA01 from four aspects: accumulation of PEP precursors, synthesis of E4P precursors, expression of core pathway enzymes, and improvement of basic resources.
Firstly, in order to increase the content of PEP, the dhaL, ptsH, and ptsI genes knocked out in E.coli SA01. Meanwhile, genes encoding glucose promoting protein (glf) and glucose kinase (glk) introduced from Zymomonas mobilis to enhance glucose uptake.
In addition, replacing the natural promoter of pykF with the growth coupled promoter PrrnC can inhibit the transcriptional activity of pykF during the stable phase, reconnecting more metabolic flows to the shikimic acid pathway, and ultimately producing strain E.coli SA02.
3. Optimization of shikimic Acid Production
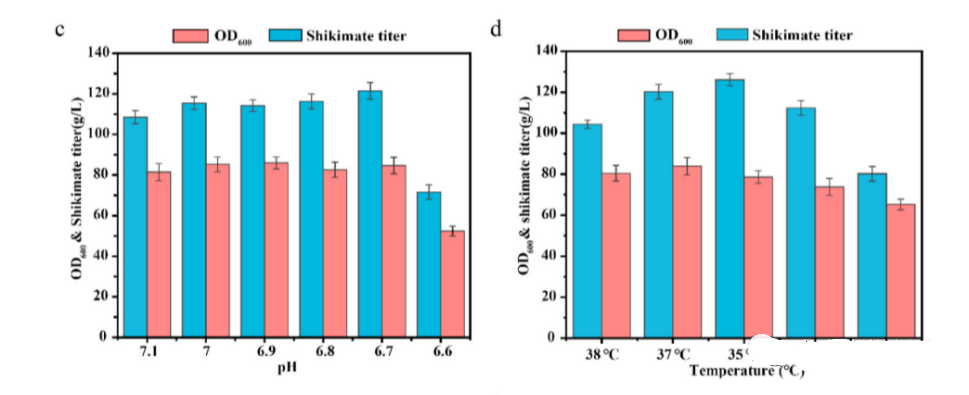
The fermentation conditions optimized, including glucose supplementation strategy, pH, and temperature, to further increase the yield of shikimic acid.
Through the use of single factor optimization experiments, it was found that when the pH and temperature of the stationary phase were controlled at 6.7 and 35 ° C, the titer, yield, and productivity of shikimic acid could be further increased to 125.9 g/L, 0.50 g/g glucose, and 2.62 g/L/h, respectively.
Enlarge it into a 30 L fermentation tank using the batch fermentation method with supplementary materials. E. Coli SA09 grows continuously from 0 to 20 hours, with an OD600 peak of 80.
During the critical production stage (20-32 hours), the average production rate of shikimic acid reached 4.44 g/L/h. At 48 hours, the titer of shikimic acid reached its maximum value of 126.4g/L, and the yield and productivity reached 0.5 g/g glucose and 2.63 g/L/h, respectively, representing the highest reported shikimic acid potency and productivity of Escherichia coli to date.
conclusion
System metabolic engineering is an effective strategy for building microbial cell factories. In this study, the goal of obtaining the required phenotype and 10 key genes for the biosynthesis of shikimic acid determined to used in computer model simulation.
In addition, using gene knockout strategies and computer protein engineering, the levels of the two main by-products quinic acid and DHS reduced by 89.1% and 52.1%, respectively.
Finally, a dynamic tolerance engineering strategy implemented by integrating high osmotic stress tolerance target genes and high concentration shikimic acid response promoters to achieve the highest shikimic acid titer and yield in Escherichia coli.